In this tutorial, you will learn how to train a Keras deep learning model to predict breast cancer in breast histology images.
Back 2012-2013 I was working for the National Institutes of Health (NIH) and the National Cancer Institute (NCI) to develop a suite of image processing and machine learning algorithms to automatically analyze breast histology images for cancer risk factors, a task that took trained pathologists hours to complete. Our work helped facilitate further advancements in breast cancer risk factor prediction
Back then deep learning was not as popular and “mainstream” as it is now. For example, the ImageNet image classification challenge had only launched in 2009 and it wasn’t until 2012 that Alex Krizhevsky, Ilya Sutskever, and Geoffrey Hinton won the competition with the now infamous AlexNet architecture.
To analyze the cellular structures in the breast histology images we were instead leveraging basic computer vision and image processing algorithms, but combining them in a novel way. These algorithms worked really well — but also required quite a bit of work to put together.
Today I thought it would be worthwhile to explore deep learning in the context of breast cancer classification.
Just last year a close family member of mine was diagnosed with cancer. And similarly, I would be willing to bet that every single reader of this blog knows someone who has had cancer at some point as well.
As deep learning researchers, practitioners, and engineers it’s important for us to gain hands-on experience applying deep learning to medical and computer vision problems — this experience can help us develop deep learning algorithms to better aid pathologists in predicting cancer.
To learn how to train a Keras deep learning model for breast cancer prediction, just keep reading!
Breast cancer classification with Keras and Deep Learning
2020-06-11 Update: This blog post is now TensorFlow 2+ compatible!
In the first part of this tutorial, we will be reviewing our breast cancer histology image dataset.
From there we’ll create a Python script to split the input dataset into three sets:
- A training set
- A validation set
- A testing set
Next, we’ll use Keras to define a Convolutional Neural Network which we’ll appropriately name “CancerNet”.
Finally, we’ll create a Python script to train CancerNet on our breast histology images.
We’ll wrap the blog post by reviewing our results.
The breast cancer histology image dataset
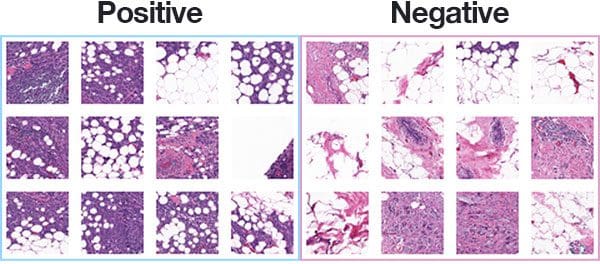
The dataset we are using for today’s post is for Invasive Ductal Carcinoma (IDC), the most common of all breast cancer.
The dataset was originally curated by Janowczyk and Madabhushi and Roa et al. but is available in public domain on Kaggle’s website.
The original dataset consisted of 162 slide images scanned at 40x.
Slide images are naturally massive (in terms of spatial dimensions), so in order to make them easier to work with, a total of 277,524 patches of 50×50 pixels were extracted, including:
- 198,738 negative examples (i.e., no breast cancer)
- 78,786 positive examples (i.e., indicating breast cancer was found in the patch)
There is clearly an imbalance in the class data with over 2x the number of negative data points than positive data points.
Each image in the dataset has a specific filename structure. An example of an image filename in the dataset can be seen below:
10253_idx5_x1351_y1101_class0.png
We can interpret this filename as:
- Patient ID: 10253_idx5
- x-coordinate of the crop: 1,351
- y-coordinate of the crop: 1,101
- Class label: 0 (0 indicates no IDC while 1 indicates IDC)
Figure 1 above shows examples of both positive and negative samples — our goal is to train a deep learning model capable of discerning the difference between the two classes.
Preparing your deep learning environment for Cancer classification
To configure your system for this tutorial, I first recommend following either of these tutorials:
Either tutorial will help you configure you system with all the necessary software for this blog post in a convenient Python virtual environment.
Please note that PyImageSearch does not recommend or support Windows for CV/DL projects.
Project structure
Go ahead and grab the “Downloads” for today’s blog post.
From there, unzip the file:
$ cd path/to/downloaded/zip $ unzip breast-cancer-classification.zip
Now that you have the files extracted, it’s time to put the dataset inside of the directory structure.
Go ahead and make the following directories:
$ cd breast-cancer-classification $ mkdir datasets $ mkdir datasets/orig
Then, head on over to Kaggle’s website and log-in. From there you can click the following link to download the dataset into your project folder:
Click here to download the data from Kaggle
Note: You will need create an account on Kaggle’s website (if you don’t already have an account) to download the dataset.
Be sure to save the .zip file in the breast-cancer-classification/datasets/orig folder.
Now head back to your terminal, navigate to the directory you just created, and unzip the data:
$ cd path/to/breast-cancer-classification/datasets/orig $ unzip archive.zip -x "IDC_regular_ps50_idx5/*"
And from there, let’s go back to the project directory and use the tree command to inspect our project structure:
$ cd ../.. $ tree --dirsfirst -L 4 . ├── datasets │ └── orig │ ├── 10253 │ │ ├── 0 │ │ └── 1 │ ├── 10254 │ │ ├── 0 │ │ └── 1 │ ├── 10255 │ │ ├── 0 │ │ └── 1 ...[omitting similar folders] │ ├── 9381 │ │ ├── 0 │ │ └── 1 │ ├── 9382 │ │ ├── 0 │ │ └── 1 │ ├── 9383 │ │ ├── 0 │ │ └── 1 │ └── 7415_10564_bundle_archive.zip ├── pyimagesearch │ ├── __init__.py │ ├── config.py │ └── cancernet.py ├── build_dataset.py ├── train_model.py └── plot.png 840 directories, 7 files
As you can see, our dataset is in the datasets/orig folder and is then broken out by faux patient ID. These images are separated into either benign (0/ ) or malignant (1/ ) directories.
Today’s pyimagesearch/ module contains our configuration and CancerNet.
Today we’ll review the following Python files in this order:
config.py: Contains our configuration that will be used by both our dataset builder and model trainer.build_dataset.py: Builds our dataset by splitting images into training, validation, and testing sets.cancernet.py: Contains our CancerNet breast cancer classification CNN.train_model.py: Responsible for training and evaluating our Keras breast cancer classification model.
The configuration file
Before we can build our dataset and train our network let’s review our configuration file.
For deep learning projects that span multiple Python files (such as this one), I like to create a single Python configuration file that stores all relevant configurations.
Let’s go ahead and take a look at config.py :
# import the necessary packages import os # initialize the path to the *original* input directory of images ORIG_INPUT_DATASET = "datasets/orig" # initialize the base path to the *new* directory that will contain # our images after computing the training and testing split BASE_PATH = "datasets/idc" # derive the training, validation, and testing directories TRAIN_PATH = os.path.sep.join([BASE_PATH, "training"]) VAL_PATH = os.path.sep.join([BASE_PATH, "validation"]) TEST_PATH = os.path.sep.join([BASE_PATH, "testing"]) # define the amount of data that will be used training TRAIN_SPLIT = 0.8 # the amount of validation data will be a percentage of the # *training* data VAL_SPLIT = 0.1
First, our configuration file contains the path to the original input dataset downloaded from Kaggle (Line 5).
From there we specify the base path to where we’re going to store our image files after creating the training, testing, and validation splits (Line 9).
Using the BASE_PATH , we derive paths to training, validation, and testing output directories (Lines 12-14).
Our TRAIN_SPLIT is the percentage of data that will be used for training (Line 17). Here I’ve set it to 80%, where the remaining 20% will be used for testing.
Of the training data, we’ll reserve some images for validation. Line 21 specifies that 10% of the training data (after we’ve split off the testing data) will be used for validation.
We’re now armed with the information required to build our breast cancer image dataset, so let’s move on.
Building the breast cancer image dataset

Our breast cancer image dataset consists of 198,783 images, each of which is 50×50 pixels.
If we were to try to load this entire dataset in memory at once we would need a little over 5.8GB.
For most modern machines, especially machines with GPUs, 5.8GB is a reasonable size; however, I’ll be making the assumption that your machine does not have that much memory.
Instead, we’ll organize our dataset on disk so we can use Keras’ ImageDataGenerator class to yield batches of images from disk without having to keep the entire dataset in memory.
But first we need to organize our dataset. Let’s build a script to do so now.
Open up the build_dataset.py file and insert the following code:
# import the necessary packages
from pyimagesearch import config
from imutils import paths
import random
import shutil
import os
# grab the paths to all input images in the original input directory
# and shuffle them
imagePaths = list(paths.list_images(config.ORIG_INPUT_DATASET))
random.seed(42)
random.shuffle(imagePaths)
# compute the training and testing split
i = int(len(imagePaths) * config.TRAIN_SPLIT)
trainPaths = imagePaths[:i]
testPaths = imagePaths[i:]
# we'll be using part of the training data for validation
i = int(len(trainPaths) * config.VAL_SPLIT)
valPaths = trainPaths[:i]
trainPaths = trainPaths[i:]
# define the datasets that we'll be building
datasets = [
("training", trainPaths, config.TRAIN_PATH),
("validation", valPaths, config.VAL_PATH),
("testing", testPaths, config.TEST_PATH)
]
This script requires that we import our config settings and paths for collecting all the image paths. We also will use random to randomly shuffle our paths, shutil to copy images, and os for joining paths and making directories. Each of these imports is listed on Lines 2-6.
To begin, we’ll grab all the imagePaths for our dataset and shuffle them (Lines 10-12).
We then compute the index of the training/testing split (Line 15). Using that index, i , our trainPaths and testPaths are constructed via slicing the imagePaths (Lines 16 and 17).
Our trainPaths are further split, this time reserving a portion for validation, valPaths (Lines 20-22).
Lines 25-29 define a list called datasets . Inside are three tuples, each with the information required to organize all of our imagePaths into training, validation, and testing data.
Let’s go ahead and loop over the datasets list now:
# loop over the datasets
for (dType, imagePaths, baseOutput) in datasets:
# show which data split we are creating
print("[INFO] building '{}' split".format(dType))
# if the output base output directory does not exist, create it
if not os.path.exists(baseOutput):
print("[INFO] 'creating {}' directory".format(baseOutput))
os.makedirs(baseOutput)
# loop over the input image paths
for inputPath in imagePaths:
# extract the filename of the input image and extract the
# class label ("0" for "negative" and "1" for "positive")
filename = inputPath.split(os.path.sep)[-1]
label = filename[-5:-4]
# build the path to the label directory
labelPath = os.path.sep.join([baseOutput, label])
# if the label output directory does not exist, create it
if not os.path.exists(labelPath):
print("[INFO] 'creating {}' directory".format(labelPath))
os.makedirs(labelPath)
# construct the path to the destination image and then copy
# the image itself
p = os.path.sep.join([labelPath, filename])
shutil.copy2(inputPath, p)
On Line 32, we define a loop over our dataset splits. Inside, we:
- Create the base output directory (Lines 37-39).
- Implement a nested loop over all input images in the current split (Line 42):
- Extract the
filenamefrom the input path (Line 45) and then extract the classlabelfrom the filename (Line 46). - Build our output
labelPathas well as create the label output directory (Lines 49-54). - And finally, copy each file into its destination (Lines 58 and 59).
- Extract the
Now that our script is coded up, go ahead and create the training, testing, and validation split directory structure by executing the following command:
$ python build_dataset.py [INFO] building 'training' split [INFO] 'creating datasets/idc/training' directory [INFO] 'creating datasets/idc/training/0' directory [INFO] 'creating datasets/idc/training/1' directory [INFO] building 'validation' split [INFO] 'creating datasets/idc/validation' directory [INFO] 'creating datasets/idc/validation/0' directory [INFO] 'creating datasets/idc/validation/1' directory [INFO] building 'testing' split [INFO] 'creating datasets/idc/testing' directory [INFO] 'creating datasets/idc/testing/0' directory [INFO] 'creating datasets/idc/testing/1' directory $ $ tree --dirsfirst --filelimit 10 . ├── datasets │ ├── idc │ │ ├── training │ │ │ ├── 0 [143065 entries] │ │ │ └── 1 [56753 entries] │ │ ├── validation │ │ | ├── 0 [15962 entries] │ │ | └── 1 [6239 entries] │ │ └── testing │ │ ├── 0 [39711 entries] │ │ └── 1 [15794 entries] │ └── orig [280 entries] ├── pyimagesearch │ ├── __init__.py │ ├── config.py │ └── cancernet.py ├── build_dataset.py ├── train_model.py └── plot.png 14 directories, 8 files
The output of our script is shown under the command.
I’ve also executed the tree command again so you can see how our dataset is now structured into our training, validation, and testing sets.
Note: I didn’t bother expanding our original datasets/orig/ structure — you can scroll up to the “Project Structure” section if you need a refresher.
CancerNet: Our breast cancer prediction CNN
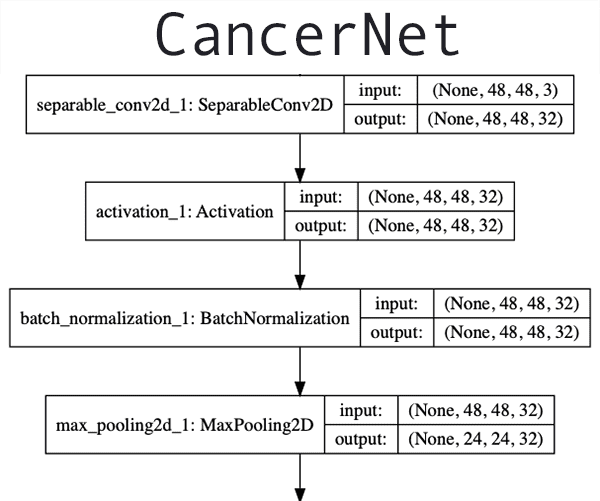
The next step is to implement the CNN architecture we are going to use for this project.
To implement the architecture I used the Keras deep learning library and designed a network appropriately named “CancerNet” which:
- Uses exclusively 3×3 CONV filters, similar to VGGNet
- Stacks multiple 3×3 CONV filters on top of each other prior to performing max-pooling (again, similar to VGGNet)
- But unlike VGGNet, uses depthwise separable convolution rather than standard convolution layers
Depthwise separable convolution is not a “new” idea in deep learning.
In fact, they were first utilized by Google Brain intern, Laurent Sifre in 2013.
Andrew Howard utilized them in 2015 when working with MobileNet.
And perhaps most notably, Francois Chollet used them in 2016-2017 when creating the famous Xception architecture.
A detailed explanation of the differences between standard convolution layers and depthwise separable convolution is outside the scope of this tutorial (for that, refer to this guide), but the gist is that depthwise separable convolution:
- Is more efficient.
- Requires less memory.
- Requires less computation.
- Can perform better than standard convolution in some situations.
I haven’t used depthwise separable convolution in any tutorials here on PyImageSearch so I thought it would be fun to play with it today.
With that said, let’s get started implementing CancerNet!
Open up the cancernet.py file and insert the following code:
# import the necessary packages from tensorflow.keras.models import Sequential from tensorflow.keras.layers import BatchNormalization from tensorflow.keras.layers import SeparableConv2D from tensorflow.keras.layers import MaxPooling2D from tensorflow.keras.layers import Activation from tensorflow.keras.layers import Flatten from tensorflow.keras.layers import Dropout from tensorflow.keras.layers import Dense from tensorflow.keras import backend as K class CancerNet: @staticmethod def build(width, height, depth, classes): # initialize the model along with the input shape to be # "channels last" and the channels dimension itself model = Sequential() inputShape = (height, width, depth) chanDim = -1 # if we are using "channels first", update the input shape # and channels dimension if K.image_data_format() == "channels_first": inputShape = (depth, height, width) chanDim = 1
Our Keras imports are listed on Lines 2-10. We’ll be using Keras’ Sequential API to build CancerNet .
An import you haven’t seen on the PyImageSearch blog is SeparableConv2D . This convolutional layer type allows for depthwise convolutions. For further details, please refer to the documentation.
The remaining imports/layer types are all discussed in both my introductory Keras Tutorial and in even greater detail inside of Deep Learning for Computer Vision with Python.
Let’s go ahead and define our CancerNet class on Line 12 and then proceed to build it on Line 14.
The build method requires four parameters:
width,height, anddepth: Here we specify the input image volume shape to our network, wheredepthis the number of color channels each image contains.classes: The number of classes our network will predict (forCancerNet, it will be2).
We go ahead and initialize our model on Line 17 and subsequently, specify our inputShape (Line 18). In the case of using TensorFlow as our backend, we’re now ready to add layers.
Other backends that specify "channels_first" require that we place the depth at the front of the inputShape and image dimensions following (Lines 23-25).
Let’s define our DEPTHWISE_CONV => RELU => POOL layers:
# CONV => RELU => POOL
model.add(SeparableConv2D(32, (3, 3), padding="same",
input_shape=inputShape))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# (CONV => RELU => POOL) * 2
model.add(SeparableConv2D(64, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(SeparableConv2D(64, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# (CONV => RELU => POOL) * 3
model.add(SeparableConv2D(128, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(SeparableConv2D(128, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(SeparableConv2D(128, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(BatchNormalization(axis=chanDim))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
Three DEPTHWISE_CONV => RELU => POOL blocks are defined here with increasing stacking and number of filters. I’ve applied BatchNormalization and Dropout as well.
Let’s append our fully connected head:
# first (and only) set of FC => RELU layers
model.add(Flatten())
model.add(Dense(256))
model.add(Activation("relu"))
model.add(BatchNormalization())
model.add(Dropout(0.5))
# softmax classifier
model.add(Dense(classes))
model.add(Activation("softmax"))
# return the constructed network architecture
return model
Our FC => RELU layers and softmax classifier make the head of the network.
The output of the softmax classifier will be the prediction percentages for each class our model will predict.
Finally, our model is returned to the training script.
Our training script
The last piece of the puzzle we need to implement is our actual training script.
Create a new file named train_model.py , open it up, and insert the following code:
# set the matplotlib backend so figures can be saved in the background
import matplotlib
matplotlib.use("Agg")
# import the necessary packages
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.callbacks import LearningRateScheduler
from tensorflow.keras.optimizers import Adagrad
from tensorflow.keras.utils import to_categorical
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from pyimagesearch.cancernet import CancerNet
from pyimagesearch import config
from imutils import paths
import matplotlib.pyplot as plt
import numpy as np
import argparse
import os
# construct the argument parser and parse the arguments
ap = argparse.ArgumentParser()
ap.add_argument("-p", "--plot", type=str, default="plot.png",
help="path to output loss/accuracy plot")
args = vars(ap.parse_args())
Our imports come from 7 places:
matplotlib: A scientific plotting package that is the de-facto standard for Python. On Line 3 we set matplotlib to use the"Agg"backend so that we’re able to save our training plots to disk.tensorflow.keras: We’ll be taking advantage of theImageDataGenerator,LearningRateScheduler,Adagradoptimizer, andutils.sklearn: From scikit-learn we’ll need its implementation of aclassification_reportand aconfusion_matrix.pyimagesearch: We’re going to be putting our newly defined CancerNet to use (training and evaluating it). We’ll also need our config to grab the paths to our three data splits. This module is not pip-installable; it is included the “Downloads” section of today’s post.imutils: I’ve made my convenience functions publicly available as a pip-installable package. We’ll be using thepathsmodule to grab paths to each of our images.numpy: The typical tool used by data scientists for numerical processing with Python.- Python: Both
argparseandosare built into Python installations. We’ll useargparseto parse a command line argument.
Let’s parse our one and only command line argument, --plot . With this argument provided in a terminal at runtime, our script will be able to dynamically accept different plot filenames. If you don’t specify a command line argument with the plot filename, a default of plot.png will be used.
Now that we’ve imported the required libraries and we’ve parsed command line arguments, let’s define training parameters including our training image paths and account for class imbalance:
# initialize our number of epochs, initial learning rate, and batch # size NUM_EPOCHS = 40 INIT_LR = 1e-2 BS = 32 # determine the total number of image paths in training, validation, # and testing directories trainPaths = list(paths.list_images(config.TRAIN_PATH)) totalTrain = len(trainPaths) totalVal = len(list(paths.list_images(config.VAL_PATH))) totalTest = len(list(paths.list_images(config.TEST_PATH))) # calculate the total number of training images in each class and # initialize a dictionary to store the class weights trainLabels = [int(p.split(os.path.sep)[-2]) for p in trainPaths] trainLabels = to_categorical(trainLabels) classTotals = trainLabels.sum(axis=0) classWeight = dict() # loop over all classes and calculate the class weight for i in range(0, len(classTotals)): classWeight[i] = classTotals.max() / classTotals[i]
Lines 28-30 define the number of training epochs, initial learning rate, and batch size.
From there, we grab our training image paths and determine the total number of images in each of the splits (Lines 34-37).
We’ll then go ahead and take steps to account for class imbalance/skew (Lines 41-48).
Let’s initialize our data augmentation object:
# initialize the training data augmentation object trainAug = ImageDataGenerator( rescale=1 / 255.0, rotation_range=20, zoom_range=0.05, width_shift_range=0.1, height_shift_range=0.1, shear_range=0.05, horizontal_flip=True, vertical_flip=True, fill_mode="nearest") # initialize the validation (and testing) data augmentation object valAug = ImageDataGenerator(rescale=1 / 255.0)
Data augmentation, a form of regularization, is important for nearly all deep learning experiments to assist with model generalization. The method purposely perturbs training examples, changing their appearance slightly, before passing them into the network for training. This partially alleviates the need to gather more training data, though more training data will rarely hurt your model.
Our data augmentation object, trainAug is initialized on Lines 51-60. As you can see, random rotations, shifts, shears, and flips will be applied to our data as it is generated. Rescaling our image pixel intensities to the range [0, 1] is handled by the trainAug generator as well as the valAug generator defined on Line 63.
Let’s initialize each of our generators now:
# initialize the training generator trainGen = trainAug.flow_from_directory( config.TRAIN_PATH, class_mode="categorical", target_size=(48, 48), color_mode="rgb", shuffle=True, batch_size=BS) # initialize the validation generator valGen = valAug.flow_from_directory( config.VAL_PATH, class_mode="categorical", target_size=(48, 48), color_mode="rgb", shuffle=False, batch_size=BS) # initialize the testing generator testGen = valAug.flow_from_directory( config.TEST_PATH, class_mode="categorical", target_size=(48, 48), color_mode="rgb", shuffle=False, batch_size=BS)
Here we initialize the training, validation, and testing generator. Each generator will provide batches of images on demand, as is denoted by the batch_size parameter.
Let’s go ahead and initialize our model and start training!
# initialize our CancerNet model and compile it model = CancerNet.build(width=48, height=48, depth=3, classes=2) opt = Adagrad(lr=INIT_LR, decay=INIT_LR / NUM_EPOCHS) model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"]) # fit the model H = model.fit( x=trainGen, steps_per_epoch=totalTrain // BS, validation_data=valGen, validation_steps=totalVal // BS, class_weight=classWeight, epochs=NUM_EPOCHS)
2020-06-11 Update: Formerly, TensorFlow/Keras required use of a method called .fit_generator in order to accomplish data augmentation. Now, the .fit method can handle data augmentation as well, making for more-consistent code. This also applies to the migration from .predict_generator to .predict (our next code block). Be sure to check out my articles about fit and fit_generator as well as data augmentation.
Our model is initialized with the Adagrad optimizer on Lines 93-95.
We then compile our model with a "binary_crossentropy" loss function (since we only have two classes of data), as well as learning rate decay (Lines 96 and 97).
Lines 100-106 initiate our training process.
After training is complete, we’ll evaluate the model on the testing data:
# reset the testing generator and then use our trained model to
# make predictions on the data
print("[INFO] evaluating network...")
testGen.reset()
predIdxs = model.predict(x=testGen, steps=(totalTest // BS) + 1)
# for each image in the testing set we need to find the index of the
# label with corresponding largest predicted probability
predIdxs = np.argmax(predIdxs, axis=1)
# show a nicely formatted classification report
print(classification_report(testGen.classes, predIdxs,
target_names=testGen.class_indices.keys()))
Line 112 makes predictions on all of our testing data (again using a generator object).
The highest prediction indices are grabbed for each sample (Line 116) and then a classification_report is printed conveniently to the terminal (Lines 119 and 120).
Let’s gather additional evaluation metrics:
# compute the confusion matrix and and use it to derive the raw
# accuracy, sensitivity, and specificity
cm = confusion_matrix(testGen.classes, predIdxs)
total = sum(sum(cm))
acc = (cm[0, 0] + cm[1, 1]) / total
sensitivity = cm[0, 0] / (cm[0, 0] + cm[0, 1])
specificity = cm[1, 1] / (cm[1, 0] + cm[1, 1])
# show the confusion matrix, accuracy, sensitivity, and specificity
print(cm)
print("acc: {:.4f}".format(acc))
print("sensitivity: {:.4f}".format(sensitivity))
print("specificity: {:.4f}".format(specificity))
Here we compute the confusion_matrix and then derive the accuracy, sensitivity , and specificity (Lines 124-128). The matrix and each of these values is then printed in our terminal (Lines 131-134).
Finally, let’s generate and store our training plot:
# plot the training loss and accuracy
N = NUM_EPOCHS
plt.style.use("ggplot")
plt.figure()
plt.plot(np.arange(0, N), H.history["loss"], label="train_loss")
plt.plot(np.arange(0, N), H.history["val_loss"], label="val_loss")
plt.plot(np.arange(0, N), H.history["accuracy"], label="train_acc")
plt.plot(np.arange(0, N), H.history["val_accuracy"], label="val_acc")
plt.title("Training Loss and Accuracy on Dataset")
plt.xlabel("Epoch #")
plt.ylabel("Loss/Accuracy")
plt.legend(loc="lower left")
plt.savefig(args["plot"])
2020-06-11 Update: In order for this plotting snippet to be TensorFlow 2+ compatible the H.history dictionary keys are updated to fully spell out “accuracy” sans “acc” (i.e., H.history["val_accuracy"] and H.history["accuracy"]). It is semi-confusing that “val” is not spelled out as “validation”; we have to learn to love and live with the API and always remember that it is a work in progress that many developers around the world contribute to.
Our training history plot consists of training/validation loss and training/validation accuracy. These are plotted over time so that we can spot over/underfitting.
Breast cancer prediction results
We’ve now implemented all the necessary Python scripts!
Let’s go ahead and train CancerNet on our breast cancer dataset.
Before continuing, ensure you have:
- Configured your deep learning environment with the necessary libraries/packages listed in the “Preparing your deep learning environment for Cancer classification” section.
- Used the “Downloads” section of this tutorial to download the source code.
- Downloaded the breast cancer dataset from Kaggle’s website.
- Unzipped the dataset and executed the
build_dataset.pyscript to create the necessary image + directory structure.
After you’ve ticked off the four items above, open up a terminal and execute the following command:
$ python train_model.py
Found 199818 images belonging to 2 classes.
Found 22201 images belonging to 2 classes.
Found 55505 images belonging to 2 classes.
Epoch 1/40
6244/6244 [==============================] - 142s 23ms/step - loss: 0.5954 - accuracy: 0.8211 - val_loss: 0.5407 - val_accuracy: 0.7796
Epoch 2/40
6244/6244 [==============================] - 135s 22ms/step - loss: 0.5520 - accuracy: 0.8333 - val_loss: 0.4786 - val_accuracy: 0.8097
Epoch 3/40
6244/6244 [==============================] - 133s 21ms/step - loss: 0.5423 - accuracy: 0.8358 - val_loss: 0.4532 - val_accuracy: 0.8202
...
Epoch 38/40
6244/6244 [==============================] - 133s 21ms/step - loss: 0.5248 - accuracy: 0.8408 - val_loss: 0.4269 - val_accuracy: 0.8300
Epoch 39/40
6244/6244 [==============================] - 133s 21ms/step - loss: 0.5254 - accuracy: 0.8415 - val_loss: 0.4199 - val_accuracy: 0.8318
Epoch 40/40
6244/6244 [==============================] - 133s 21ms/step - loss: 0.5244 - accuracy: 0.8422 - val_loss: 0.4219 - val_accuracy: 0.8314
[INFO] evaluating network...
precision recall f1-score support
0 0.93 0.83 0.88 39853
1 0.66 0.85 0.75 15652
accuracy 0.84 55505
macro avg 0.80 0.84 0.81 55505
weighted avg 0.86 0.84 0.84 55505
[[33107 6746]
[ 2303 13349]]
acc: 0.8370
sensitivity: 0.8307
specificity: 0.8529

Looking at our output you can see that our model achieved ~83% accuracy; however, that raw accuracy is heavily weighted by the fact that we classified “benign/no cancer” correctly 93% of the time.
To understand our model’s performance at a deeper level we compute the sensitivity and the specificity.
Our sensitivity measures the proportion of the true positives that were also predicted as positive (83.07%).
Conversely, specificity measures our true negatives (85.29%).
We need to be really careful with our false negative here — we don’t want to classify someone as “No cancer” when they are in fact “Cancer positive”.
Our false positive rate is also important — we don’t want to mistakenly classify someone as “Cancer positive” and then subject them to painful, expensive, and invasive treatments when they don’t actually need them.
There is always a balance between sensitivity and specificity that a machine learning/deep learning engineer and practitioner must manage, but when it comes to deep learning and healthcare/health treatment, that balance becomes extremely important.
For more information on sensitivity, specificity, true positives, false negatives, true negatives, and false positives, refer to this guide.
What's next? We recommend PyImageSearch University.
84 total classes • 114+ hours of on-demand code walkthrough videos • Last updated: February 2024
★★★★★ 4.84 (128 Ratings) • 16,000+ Students Enrolled
I strongly believe that if you had the right teacher you could master computer vision and deep learning.
Do you think learning computer vision and deep learning has to be time-consuming, overwhelming, and complicated? Or has to involve complex mathematics and equations? Or requires a degree in computer science?
That’s not the case.
All you need to master computer vision and deep learning is for someone to explain things to you in simple, intuitive terms. And that’s exactly what I do. My mission is to change education and how complex Artificial Intelligence topics are taught.
If you're serious about learning computer vision, your next stop should be PyImageSearch University, the most comprehensive computer vision, deep learning, and OpenCV course online today. Here you’ll learn how to successfully and confidently apply computer vision to your work, research, and projects. Join me in computer vision mastery.
Inside PyImageSearch University you'll find:
- ✓ 84 courses on essential computer vision, deep learning, and OpenCV topics
- ✓ 84 Certificates of Completion
- ✓ 114+ hours of on-demand video
- ✓ Brand new courses released regularly, ensuring you can keep up with state-of-the-art techniques
- ✓ Pre-configured Jupyter Notebooks in Google Colab
- ✓ Run all code examples in your web browser — works on Windows, macOS, and Linux (no dev environment configuration required!)
- ✓ Access to centralized code repos for all 536+ tutorials on PyImageSearch
- ✓ Easy one-click downloads for code, datasets, pre-trained models, etc.
- ✓ Access on mobile, laptop, desktop, etc.
Summary
In this tutorial, you learned how to use the Keras deep learning library to train a Convolutional Neural Network for breast cancer classification.
To accomplish this task, we leveraged a breast cancer histology image dataset curated by Janowczyk and Madabhushi and Roa et al.
The histology images themselves are massive (in terms of image size on disk and spatial dimensions when loaded into memory), so in order to make the images easier for us to work with them, Paul Mooney, part of the community advocacy team at Kaggle, converted the dataset to 50×50 pixel image patches and then uploaded the modified dataset directly to the Kaggle dataset archive.
A total of 277,524 images belonging to two classes are included in the dataset:
- Positive (+): 78,786
- Negative (-): 198,738
Here we can see there is a class imbalance in the data with over 2x more negative samples than positive samples.
The class imbalance, along with the challenging nature of the dataset, lead to us obtaining ~83% classification accuracy, ~83% sensitivity, and ~85% specificity.
I invite you to use this code as a template for starting your own breast cancer classification experiments.
To download the source code to this post, and be notified when future tutorials are published here on PyImageSearch, just enter your email address in the form below!
Download the Source Code and FREE 17-page Resource Guide
Enter your email address below to get a .zip of the code and a FREE 17-page Resource Guide on Computer Vision, OpenCV, and Deep Learning. Inside you'll find my hand-picked tutorials, books, courses, and libraries to help you master CV and DL!


